Product Line
ReneGENE: Accurate and accelerated solution for secondary analysis of genome
- Proprietary core technology, currently offered on HES-HPCâ„¢ (from Aldec) as AccuRA, for accurate, and ultra-fast big data mapping and alignment of DNA short-reads from the Next Generation Sequencing (NGS) platforms, with full coverage of the genome (million to billion bases long), including repeat regions.
- Fully streaming, multithreaded, parallel dynamic programming in hardware eliminates memory bottleneck and storage issues.
- Reduces computing and I/O burden on the host significantly.
- Scalable and massively parallel computing and data pipeline.
- Achieves short read mapping in minimum deterministic time.
Service Line
ReneGENE is in the process of being developed into a cloud-based service.
Solutions
AccuRA: core mapping and alignment technology implemented on reconfigurable hardware platform
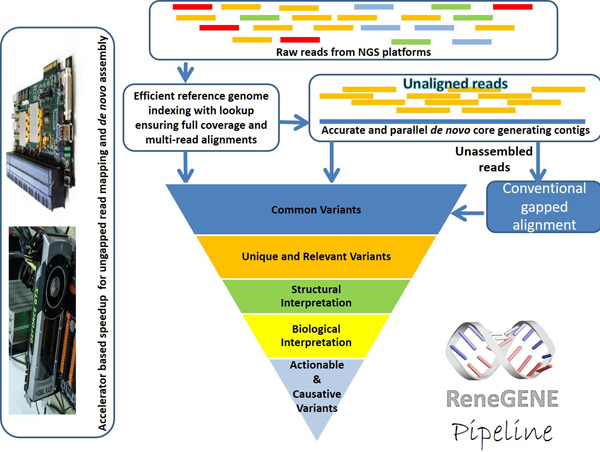
Classified as a big data problem, Short Read Mapping (SRM) within the Next Generation Sequencing (NGS) pipeline presents profound technical and computing challenges. Existing solutions handle the high volume of data leveraging heuristics, to claim notable performance standards. The results from SRM leave a huge impact across fields, including medical diagnostics and drug discovery. In this context, we need precise, affordable, reliable and actionable results from SRM, to support any application, with uncompromised accuracy and performance. AccuRA, is the technology implementation of ReneGENE on reconfigurable hardware platform, using a massively parallel, scalable accelerator for accurate alignment of short reads. Supplemented with multithreaded firmware architecture, AccuRA precisely aligns short reads, at a fine-grained single nucleotide resolution, and offers full coverage of the genome. AccuRA is a fully streaming hardware solution using dynamic programming for SRM in the minimum possible and deterministic time. AccuRA also scales well at multiple levels of design granularity, while successfully aligning genomes of various sizes, ranging from small archeal, bacterial, fungal genomes, to the large mammalian human genome.
GMAccS
GMAccS is a GPU based OpenCL implementation of ReneGENE that is scalable and massively parallel archetype for accelerating Short Read Mapping (SRM) within Next Generation Sequencing (NGS) data analysis pipeline. Existing solutions for accelerating SRM provide notable performance, while leveraging heuristics. The results from SRM greatly impact the clinical decision making process in therapeutics and drug discovery and demand great accuracy, reliability and actionable results from SRM. In this context, GMAccS performs accurate alignment of short reads, at a fine grained single nucleotide resolution, by delivering single character accuracy in string matching. GMAccS offers full coverage of the human genome, by effectively handling repeat regions and redundant reads without heuristics. GMAccS successfully scales at multiple levels of granularity, while aligning the smaller archeal genomes, to the much larger mammalian human genomes.